
Contents


|
Programme
Contents 

|
Potent blockers of Kv1.3 potassium channel are crucial in the activation of human effector memory T cells (TEM); selective blockers constitute valuable therapeutic leads for the treatment of autoimmune diseases mediated by TEM cells. Computational simulation at the molecular level is a powerful tool in understanding electrophysiological experiments performed on wild type and mutant channel. Here we go for molecular dynamics (MD) simulation on complex Kv1.3-Shk (sea anemone toxin) to measure the potency of Shk to channel Kv1.3.
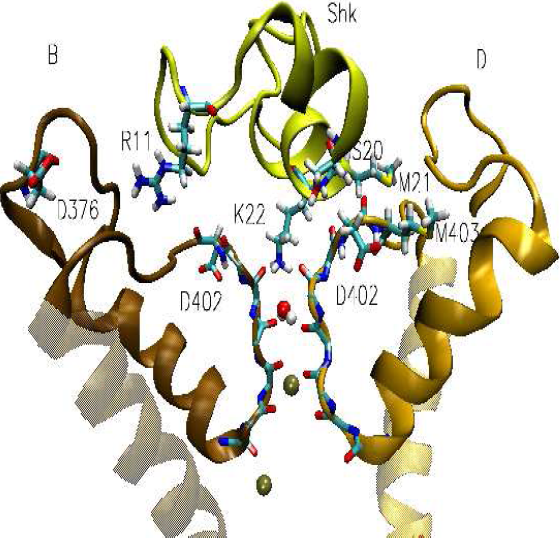
Side view of Shk in complex with Kv1.3 potassium channel. Shk backbone is shown in yellow and the side chains of R11, M21, K22 and S20 residues involve in binding are explicitly shown. Two of the four monomers in Kv1.3 (B and D) are shown clearly.
Based on crystal structure of voltage-gated potassium channel Kv1.2 we have constructed a homology model of Kv1.3. Haddock has been used for the exploration of the conformational space and the determination of ligand /protein contacts. The potassium ion channel axis provides a natural reaction coordinate for unbinding of a sea anemone toxin (Shk). Along this coordinate we measure the binding energy of toxin with the channel. The computational results have an excellent platform due to a comprehensive collection of physiological data available to test the outcomes. We have run the MD simulation up to 10 ns for the channel-toxin complex in a solvated lipid bilayer environment. This confirmed that the toxin and the pore region of the channel are flexible in the binding. The last 5 ns is considered as the production time. Recognition residues and interaction contacts for the binding were identified during this time of simulation. Lys-22 goes far inside the channel. Among Kv1 family Kv1.3 has His-404 above the selectivity filter. His404 makes a hydrogen bond with Asp402 of the next monomer in a pattern of His(A)-Asp(B), His(B)-Asp(C), His(C)-Asp(D) and His(D)-Asp(A). This His(404)/Asp(402) pair does not allow Asp402 side chain to make any interaction with toxin. Arg11 comes closer to residues Asp376(B), His404(B), Ser379(B) and Asp402(B). Met21/Met403(D), His19/Asp402(D), Tyr23/Gly401(C) are the interacting residue pairs in the Shk-Kv1.3 complex. This is an agreement with the experimental (mutant cycle analysis) result (Lanigan et al., 2002). To measure the potency of Shk we calculate the potential of mean force (PMF) of its unbinding from the channel. Weighted histogram analysis method (WHAM) is used to calculate PMF from Umbrella sampling data. The outcome PMF from the umbrella sampling simulation differ 1 kcal/mol from the experimental measurement. The consistency between the result of the simulations and the experimental data indicate that our three-dimensional models of the toxin-channel complex are reasonable. So our model can be used as a guide for future biological studies, such as the rational design of selective blocking agents of the Kv1.3 channel and mutation cycle analysis in toxins to increase selectivity and potency towards the channel. Thereby we are in search for more potent mutants of Shk.
Lanigan MD, Kalman K, Lefievre Y, Pennington MW, Chandy KG, Norton RS. (2002) Biochemistry, 41: 11963-11971.