Introduction. Genome research is a fast growing and computationally demanding research domain, characterized by bottlenecks in expertise which are compounded by lack of accessible analysis and visualization infrastructure. Australia has developed significant high-performance computing (HPC) resources as part of the National Computing Infrastructure (NCI, see http://nf.nci.org.au/), NeCTAR and other programs. Some challenges in genomics, however, are different to many other research endeavours that are solved through HPC. Genome research is a data-intensive form of discovery, requiring the amassing, indexing and analysis of vast amounts of data, and its comparison to large public catalogues of genomic knowledge. Typical genomics analyses will involve multiple stages of data transformation using existing tools in an environment of rapidly evolving best practice and current knowledge.
Fortunately, many computational problems in genomics have been solved. For example, visualization of genomic data is available with the UCSC (Kent et al., 2002), GBrowse (http://gmod.org/wiki/GBrowse/), IGV (Nicol et al., 2009), and other genome browsers. Using workflow platforms such as Galaxy (Giardine et al., 2005) or GenePattern (Reich et al., 2006), biologists can develop scientific workflows (or ‘pipelines’) using a web browser and launch tasks on HPC clusters. Many groups in Australia do not have simple access to these tools and data resources, however. They are complicated to install and customise, require dedicated compute resources and data stores, and typically involve a high level of ongoing maintenance to keep the software, data and hardware current, which in turn requires significant expertise in software development, system administration, hardware and networking, as well as access to hardware resources and data-centres. Further, even with access to current infrastructure, understanding and applying best practice in genomic informatics is non-trivial, typically requiring considerable expertise, up-to-date resources, advice and support and community participation.
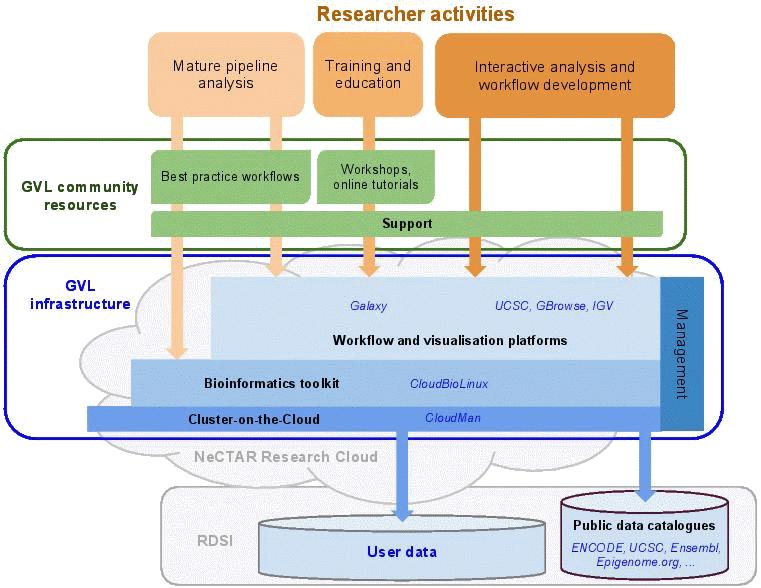
The Genomics Virtual Laboratory (GVL) has been established to provide these tools and data to genome researchers to enable collaboration nation-wide in order to ensure Australian competitiveness and leadership now and into the future.

The GVL is a collaborative initiative aiming to connect genome researchers with massive datasets, sophisticated analysis and visualization tools, and large-scale computational and storage infrastructure, supported by federal funding from the NeCTAR program plus co-investment from a number of research institutes. Essentially the GVL is a combination of scalable genomics infrastructure, analysis platforms, resources, and support (Figure 1) which will be represented at multiple physical nodes. Researchers can access one of the centrally managed nodes of the GVL to directly perform genomic analyses and visualisation, with large resource/support allocations available through subscriptions; alternatively research groups or institutes may instantiate, manage and tailor their own workflow platform on the same public cloud infrastructure, on resources reserved through the NeCTAR and RDSI resource allocation processes.
Project progress. Development of the GVL is funded under the NeCTAR Virtual Laboratories program (available from https://nectar.org.au/virtual-laboratories-0), with the infrastructure being implemented nationally in stages: in 2012 prototype nodes are being developed at UQ and the University of Melbourne, and in 2013 and beyond these facilities will be developed at additional Research Cloud nodes around Australia and moved into production. The GVL is designed to scale to multiple locations and arbitrary cluster sizes on the Research Cloud through the CloudMan platform (Afgan et al., 2010), and will be supported by comprehensive training courses, outreach programs and end-user support for subscribers.
At this stage of development, the GVL comprises: a prototype workflow management system based on the Galaxy framework (Giardine et al., 2005), a bioinformatics toolkit (for command-line users), and a visualization service based on the UCSC Genome Browser, all implemented on the NeCTAR Research Cloud (see link above); and a developing set of tutorials and exemplar workflows targeted at common high throughput genomics tasks (see Figure 1 for an illustration). New GVL instances can be implemented on the Research Cloud as required, and new instances can be tailored to specific requirements of individual research groups or institutes as required.
Further development will add a number of community-advocated workflows which can be used as templates plus extensive training and education materials, genomic data visualisation systems, local copies of important national and international reference datasets, and a science collaboration framework.
Afgan E, Baker D, Coraor N, Chapman B, Nekrutenko A, Taylor J. (2010) Galaxy CloudMan: delivering cloud compute clusters. BMC Bioinformatics 11(Suppl 12): S4.
Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A. (2005) Galaxy: A platform for interactive large-scale genome analysis. Genome Research 15: 1451-1455.
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. (2002) The human genome browser at UCSC. Genome Research 12: 996-1006.
Nicol JW, Helt GA, Blanchard SG Jr, Raja A, Loraine AE. (2009) The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25: 2730-2731.
Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. (2006) GenePattern 2.0. Nature Genetics 38: 500-501.